Chapter 4 Transfer cell annotations
4.1 Transfer cell annotations from the Allen MTG reference to the AD slides.
First step is to identify and transfer gene anchors between the Allen MTG reference and the AD slides.
We need to add SCT normalization to the Allen MTG reference, and then find the anchors. Since SCTransform requires the counts slot to be filled, we need to copy the data slot to the counts slot.
Let’s as well downlsample the Allen MTG reference to 10% of the cells, to speed up the process and go easy on the RAM consumption.
allen_MTG_reference = scMiko::downsampleSeurat(allen_MTG_reference, subsample.factor = 0.1)
allen_MTG_reference[["RNA"]]$counts = allen_MTG_reference[["RNA"]]$data
allen_MTG_reference = Seurat::SCTransform(allen_MTG_reference, verbose = TRUE)
gc()
# convert ENSEMBL to Symbols in Allen MTG reference
library("org.Hs.eg.db")
gene_list = AnnotationDbi::mapIds(
org.Hs.eg.db,
keys = rownames(allen_MTG_reference[["RNA"]]$counts),
keytype = "ENSEMBL",
column = "SYMBOL",
multiVals = "first"
)
allen_MTG_reference = scMiko::ens2sym.so(allen_MTG_reference, gene_list)
# drop NAs
gene_list = gene_list[!is.na(gene_list)]
allen_MTG_reference = subset(allen_MTG_reference, features = gene_list)
# fix rownames in the meta.features slot
rownames(allen_MTG_reference[["SCT"]]@meta.features) = rownames(allen_MTG_reference[["SCT"]]@data)Now that we have reformatted the Allen MTG reference, we finally proceed with the transfer of cell annotations.
# get anchors
anchors = Seurat::FindTransferAnchors(
reference = allen_MTG_reference,
query = ST011,
normalization.method = "SCT",
recompute.residuals = FALSE
)
# transfer cell_type annotations to ST011 slides
predictions_assay = Seurat::TransferData(
anchorset = anchors,
refdata = allen_MTG_reference$cell_type,
prediction.assay = TRUE,
weight.reduction = ST011[["pca"]],
dims = 1:30
)
# add predictions to ST011 as a new slot
ST011[["predictions"]] = predictions_assay
Seurat::DefaultAssay(ST011) = "predictions"
# add predictions to the metadata
predictions_id = Seurat::TransferData(
anchorset = anchors,
refdata = allen_MTG_reference$cell_type,
prediction.assay = FALSE,
weight.reduction = ST011[["pca"]],
dims = 1:30
)
ST011$predicted_id = predictions_id$predicted_id
ST011[[]]$predicted_id = predictions_id$predicted_idLet’s now check the voxels predicted to contains astrocytes, oligodendrocytes and fibroblasts in the AD slides compared to the control slides.
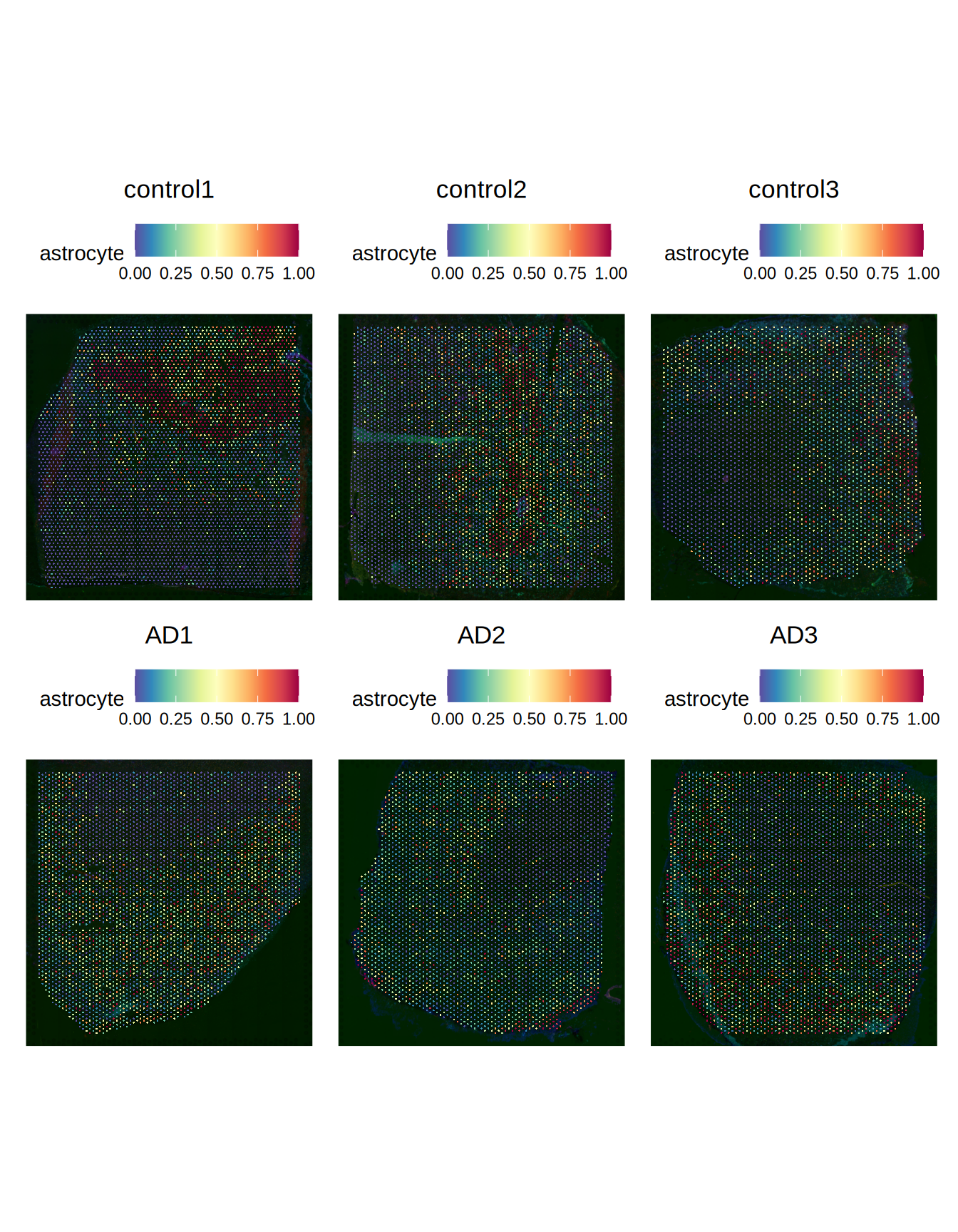
Figure 4.1: Predicted astrocytes
SpatialFeaturePlot(
ST011,
features = "oligodendrocyte",
pt.size.factor = 1500,
ncol = 3,
crop = TRUE
)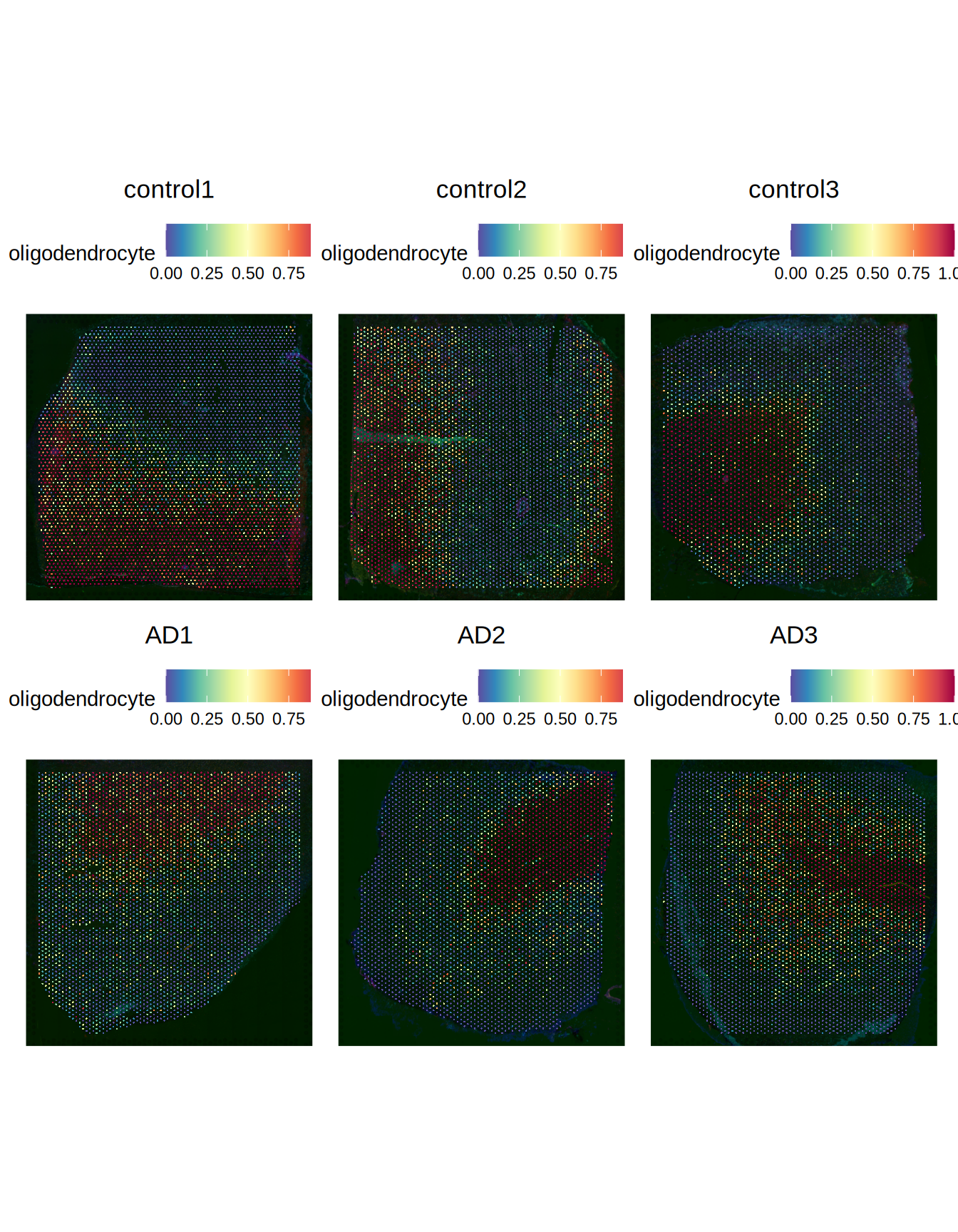
Figure 4.2: Predicted oligodendrocytes
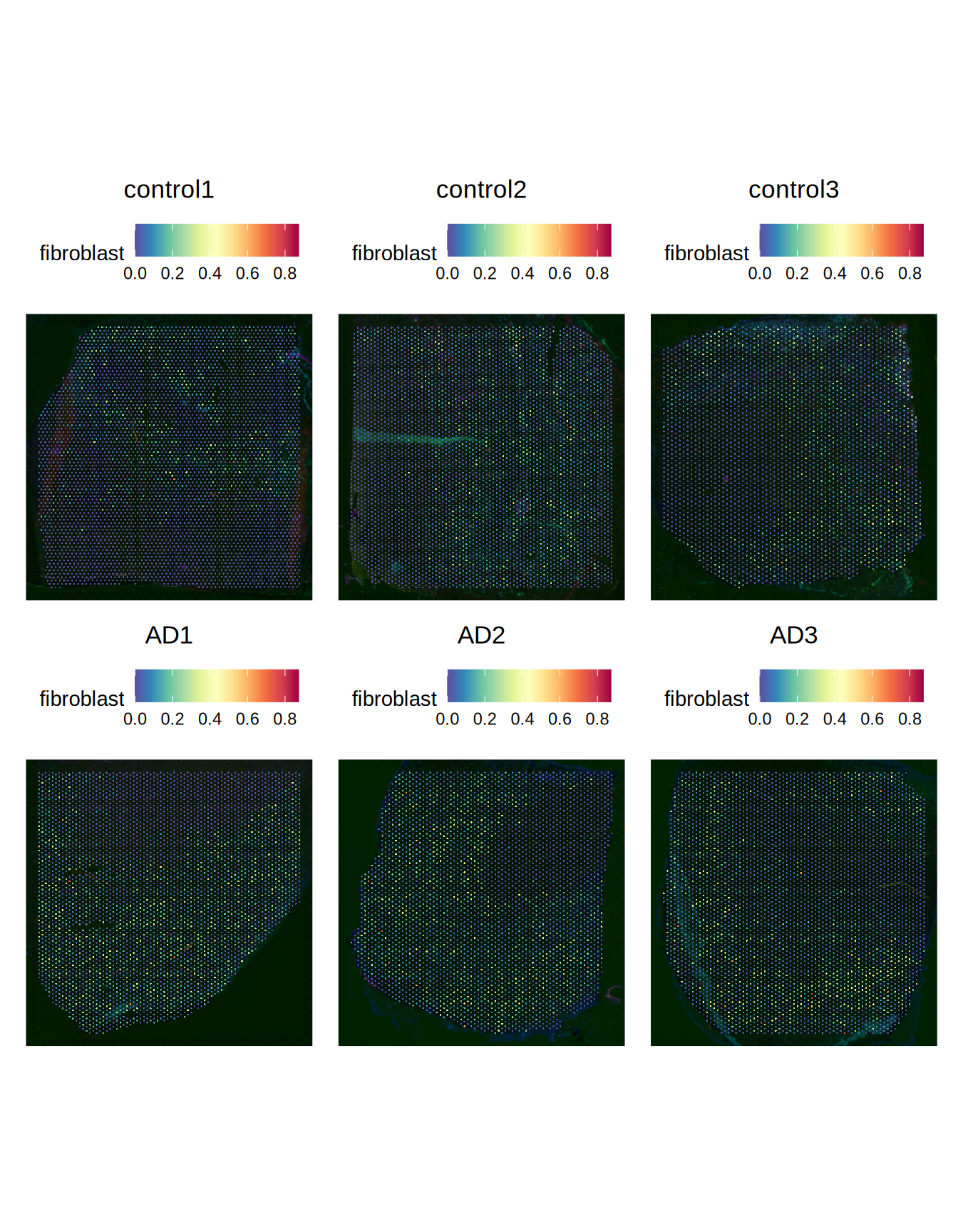
Figure 4.3: Predicted fibrolasts
We can notice a decrease of astrocytes and and increase in fibroblasts in the AD slides when compared to the control slides.