8 Figure 2
8.1 Figure 2 code
8.1.1 Panel A code
library(ape)
library(ggtree)
library(ggplot2)
library(RColorBrewer)
# Read tree in newick format using ape
kveik_tree <- read.tree('DendroExport_densitybased.txt')
# Identify tip labels that start with "AR" or "AP" as we dont have phenotype data for culture A
#tips_to_drop <- kveik_tree$tip.label[grep("^(AR|AP)", kveik_tree$tip.label)]
# rename A to AB
kveik_tree$tip.label <- sub("^AR([0-9]+)$", "ABR\\1", kveik_tree$tip.label)
kveik_tree$tip.label <- sub("^AP([0-9]+)$", "ABP\\1", kveik_tree$tip.label)
# add 'R' strains to be swapped out to drop list
tips_to_drop <- c(
'21R40',
'17R20',
'45R38',
'28R31',
'28R1')
# Drop the identified tips
kveik_tree <- drop.tip(kveik_tree, tips_to_drop)
# list strains to relabel
label_changes <- c("21P1" = "21R40",
"17P5" = "17R20",
"45P5" = "45R38",
"28P1" = "28R31",
"28P6" = "28R1")
# Replace the specified tip labels
kveik_tree$tip.label <- sapply(kveik_tree$tip.label, function(label) {
if (label %in% names(label_changes)) {
return(label_changes[label])
} else {
return(label)
}
})
# Identify tip labels that are "P" strains
tips_to_drop <- grep("P", kveik_tree$tip.label, value = TRUE)
# Drop the P strains
kveik_tree <- drop.tip(kveik_tree, tips_to_drop)
# extract group info from tip labels (label format = culture R picking number e.g. 1R1, want to extract only culture number)
gpinfo <- split(kveik_tree$tip.label, gsub("[RPb]\\w+", "", kveik_tree$tip.label))
# assign group info to tree
kveik_tree <- groupOTU(kveik_tree, gpinfo)
# plot tree, circular cladogram with colors by group
p <- ggtree(kveik_tree, aes(color = group), size = 0.25, layout = 'circular') +
geom_tiplab2(aes(angle = angle), size = 0.5) +
labs(title = 'Interdelta band clustering', color = 'Farmhouse Culture') +
theme(plot.title = element_text(hjust = 0.5, size = 20),
legend.title = element_text(size = 15),
legend.text = element_text(size = 12)) +
guides(color = guide_legend(override.aes = list(label = "", size = 1)))
# read in info file
heatmapData <- read.csv('heatmap data - swaped P out.csv', row.names = 1)
rn <- rownames(heatmapData)
heatmapData <- heatmapData[, c(3,4)] # select which columns to display here
heatmapData <- as.data.frame(sapply(heatmapData, as.character))
rownames(heatmapData) <- rn
heatmap.colours <- c('0' = '#FFFFFF',
'1' = '#0571B0',
'2' = '#92C5DE',
'3' = '#018571',
'4' = '#80CDC1',
'5' = '#FFDA00',
'6' = '#FBA01D',
'7' = '#A6611A',
'8' = '#FF0000')
pdf(file = 'cladogram updated 2.pdf', width = 10, height = 8)
gheatmap(p, heatmapData, offset = 1, color=NA, width = 0.1,
colnames = FALSE) +
scale_fill_manual(values=heatmap.colours, breaks=c(1:7,0,8), labels = c('North-West Norway', 'South-West Norway', 'Central-Eastern Norway', 'South-Eastern Norway','Latvia', 'Lithuania','Russia', '', 'sequenced'), aes(legend_title = 'Location'))
dev.off()
# ###########################################
# # label nodes
# p1 <- p + geom_text(aes(label=node), hjust=-.3)
# pdf(file = 'nodes.pdf', width = 30, height = 30)
# p1
# dev.off()
# 8.1.2 Merge
panel_a = ggplot2::ggplot() + ggplot2::annotation_custom(
grid::rasterGrob(
magick::image_read("data/p02-02/cladogram updated 2.pdf"),
width = ggplot2::unit(1,"npc"),
height = ggplot2::unit(1,"npc")),
-Inf, Inf, -Inf, Inf)
panel_b = ggplot2::ggplot() + ggplot2::annotation_custom(
grid::rasterGrob(
magick::image_read("data/p02-02/graph2.pdf"),
width = ggplot2::unit(1,"npc"),
height = ggplot2::unit(1,"npc")),
-Inf, Inf, -Inf, Inf)
final_plot = cowplot::plot_grid(
panel_a,
panel_b,
nrow = 2,
rel_heights = c(3, 1),
labels = c("A", "B")
)8.2 Figure 2 plot
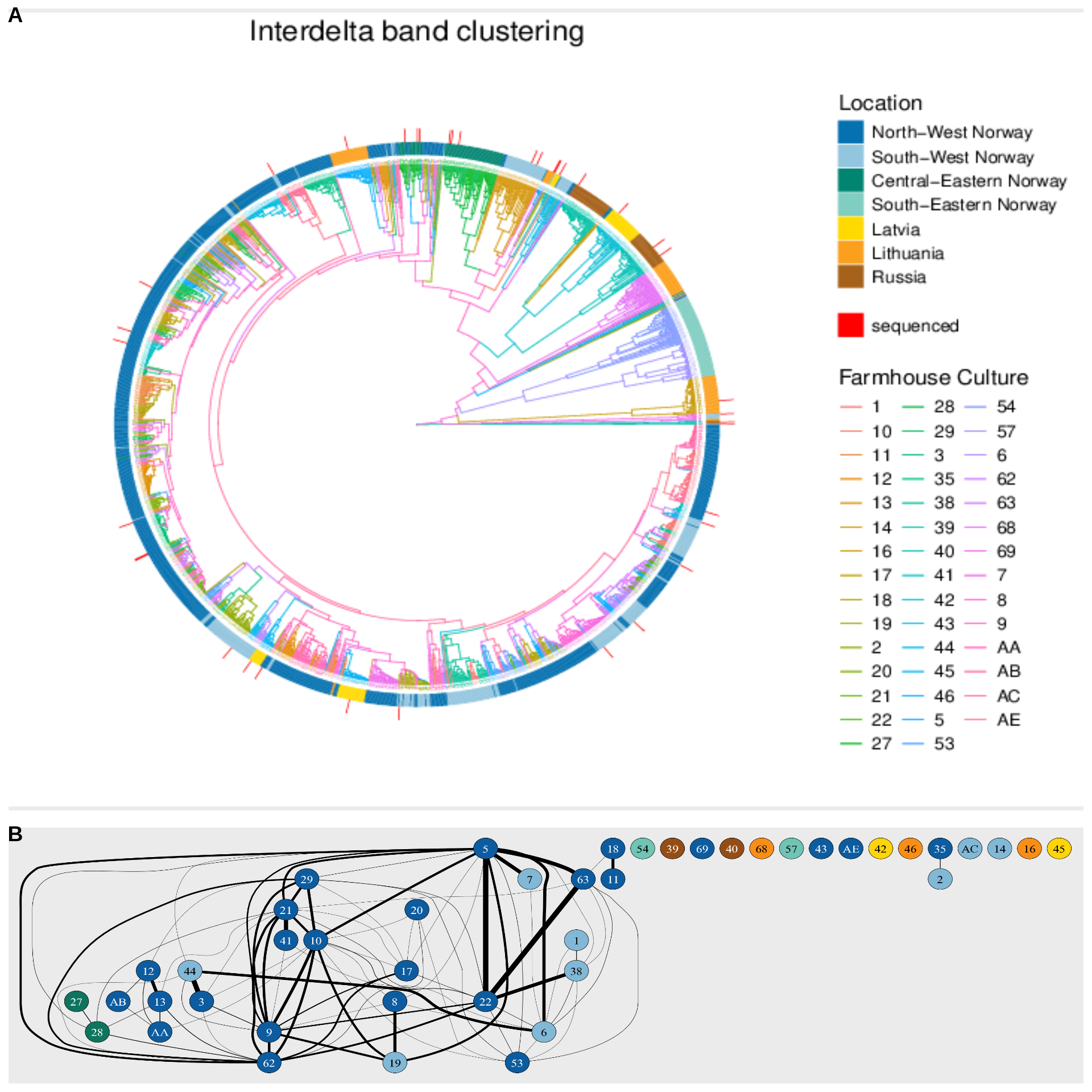
8.3 Session Information
Note
R version 4.3.3 (2024-02-29)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 24.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.12.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=en_US.UTF-8 LC_MONETARY=C LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
time zone: Europe/Brussels
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] RColorBrewer_1.1-3 ggtext_0.1.2 ggrepel_0.9.6 ggplot2_3.5.2
[5] forcats_1.0.0 dplyr_1.1.4 cowplot_1.2.0
loaded via a namespace (and not attached):
[1] gtable_0.3.6 jsonlite_2.0.0 compiler_4.3.3 tidyselect_1.2.1
[5] Rcpp_1.1.0 xml2_1.3.8 magick_2.8.7 dichromat_2.0-0.1
[9] scales_1.4.0 yaml_2.3.10 fastmap_1.2.0 R6_2.6.1
[13] labeling_0.4.3 generics_0.1.4 knitr_1.50 htmlwidgets_1.6.4
[17] tibble_3.3.0 pillar_1.11.0 rlang_1.1.6 xfun_0.52
[21] cli_3.6.5 withr_3.0.2 magrittr_2.0.3 digest_0.6.37
[25] grid_4.3.3 gridtext_0.1.5 lifecycle_1.0.4 vctrs_0.6.5
[29] evaluate_1.0.4 glue_1.8.0 farver_2.1.2 rmarkdown_2.29
[33] tools_4.3.3 pkgconfig_2.0.3 htmltools_0.5.8.1