18 Figure S6
18.1 Figure S6 code
18.1.1 Panel A code
#==============================================================================#
# 1 - k-mer frequencies ####
#------------------------------------------------------------------------------#
## 1.1 - Import and reformat k-mer freq ####
# mapping swap_yeasts
to_swap = data.frame(
old_name = c("21P1", "17P5", "45P5", "28P1", "28P6"),
new_name = c("21R40", "17R20", "45R38", "28R31", "28R1")
)
# load the tree file to make dendrogram
tree = read.tree("./data/p02-12/genetrees.output.BS.best.2025.tre")
# Define the specific samples to keep
selected_samples = c(
"1R16", "2R23", "3R11", "6R15", "k7R25", "7R7", "8R19", "9R40", "14R30", "14R6",
"16R23", "16R37", "17P5", "19R18", "21P1", "21R38", "27R17", "28P1", "28P6", "28R21",
"28R33", "28R8", "38R16", "39R20", "40R1", "40R14", "40R20", "41R10", "41R15", "42R20",
"42R31", "44R32", "44R7", "45P5", "45R11", "46R12", "46R37", "Hornindal1", "Hornindal2",
"Laerdal2", "Muri" , "SortdalEbbe1", "Voss1", "Granvin1", "Skud"
)
# Prune the tree to keep only the selected samples
pruned_tree = keep.tip(tree, selected_samples)
# Reroot the pruned tree with "Skud" as outgroup
pruned_tree = root(pruned_tree, outgroup = "Skud", resolve.root = FALSE)
# Prune the pruned tree to remove Skud
pruned_tree = drop.tip(pruned_tree, "Skud")
# dendrogram plot
p_tree = # Plot the pruned tree
ggtree(pruned_tree) +
theme(plot.margin = margin(10, 10, 10, 10)) +
xlim(0, 15)
# extract tip order
tip_order = p_tree$data %>%
filter(isTip) %>% # Select only tip labels
arrange(y) %>% # Arrange by y-axis position
pull(label)
for(k in 1:nrow(to_swap)){
tip_order = stringr::str_replace_all(
tip_order,
to_swap[k, "old_name"],
to_swap[k, "new_name"]
)
}
# import CNV matrix
kmer41freq = read.delim("./data/p02-12/Vikings.species.kmer41freq.mod.txt", header = FALSE)
colnames(kmer41freq) = c("strain", "type", "kmer", "freq")
for(k in 1:nrow(to_swap)){
kmer41freq$strain = stringr::str_replace_all(
kmer41freq$strain,
to_swap[k, "old_name"],
to_swap[k, "new_name"]
)
}
kmer41freq$strain = factor(kmer41freq$strain, levels = rev(tip_order))
kmer41freq$peak = ifelse(
kmer41freq$strain %in% c("Muri", "k7R25"), "one_peak",
ifelse(
kmer41freq$strain %in% c("38R16", "16R23", "39R20", "40R14"), "two_peaks",
ifelse(
kmer41freq$strain %in% c(
"41R10", "21R38", "21P1", "28P1", "28P6", "28R21", "28R33", "28R8",
"46R12", "46R37", "16R37", "40R1", "40R20"
), "three_peaks",
ifelse(kmer41freq$strain == "44R32", "five_peaks", "four_peaks")
)
)
)
kmer41freq$peak = factor(
kmer41freq$peak,
levels = c("one_peak", "two_peaks", "three_peaks", "four_peaks", "five_peaks")
)
#------------------------------------------------------------------------------#
## 3.2 - Plot ####
# set color labels
col_label = c("#0571B0", rep("#92C5DE", 5), "#0571B0", rep("#92C5DE", 3),
rep("#0571B0", 9), rep("#92C5DE", 4), rep("#FFDA00", 4),
rep("#FBA01D", 3), rep("#008470", 6), rep("grey75", 6),
"#A6611A", "#FBA01D")
# plot kmer distributions
p_kmer = ggplot(kmer41freq, aes(x = kmer, y = freq)) +
geom_line(aes(colour = peak), linewidth = 2) +
scale_colour_manual(values = c("tomato4", "tomato2", "skyblue1", "skyblue3", "skyblue4")) +
coord_cartesian(expand = FALSE) +
annotation_custom(grid::linesGrob(y = c(0, 0), gp = grid::gpar(lwd = 3))) +
ggh4x::facet_grid2(strain ~ type,
scales = "free",
independent = "x",
switch = "both",
strip = strip_themed(
background_y = ggh4x::elem_list_rect(fill = col_label),
text_y = ggh4x::elem_list_text(color = c(
rep("black", 36), rep("grey50", 6), rep("black", 2)
))
)) +
theme(plot.title = element_blank(),
axis.title = element_blank(),
axis.ticks = element_blank(),
axis.text = element_blank(),
legend.position = "none",
panel.background = element_blank(),
panel.border = element_blank(),
panel.spacing = unit(0, "lines"),
strip.background = element_blank(),
strip.text.x = element_text(size = 8),
strip.text.y.left = element_text(size = 8, angle = 0, face = "bold"))
#==============================================================================#
# 2 - CNVs ####
#------------------------------------------------------------------------------#
## 2.1 - Import and reformat coverage ####
# import CNV matrix
CNV_table = read.delim("./data/p02-12/Vikings.CNVsmerged.all.tab", header = TRUE)
CNV_table$CNV = as.factor(CNV_table$CNV)
# import dataset
file_list = list.files(
path = "./data/p02-12/01_start_bed/",
pattern = "1kb_cov.bed",
full.names = TRUE,
ignore.case = FALSE
)
# declare average df
average_table = data.frame(
strain = character(),
average = double(),
max = double(),
sd = double(),
stringsAsFactors = FALSE
)
# declare cov df
coverage_table = data.frame(
strain = character(),
chr = character(),
start = integer(),
stop = integer(),
cov = double(),
stringsAsFactors = FALSE
)
# import coverage counts per strain
for(file_to_import in file_list){
tmp_table = read.delim(file_to_import, header = FALSE)
colnames(tmp_table) = c("strain", "chr", "start", "stop", "cov")
tmp_table = tmp_table[which(tmp_table$chr != "ref|NC_001224|"), ]
strain_name = stringr::str_remove(file_to_import, ".align.sort.1kb_cov.bed")
strain_name = stringr::str_remove(strain_name, "./data/p02-12/01_start_bed//")
average_table = rbind(
average_table,
data.frame(
"strain" = strain_name,
"average" = mean(tmp_table$cov),
"max" = max(tmp_table$cov),
"sd" = sd(tmp_table$cov)
)
)
tmp_table = tmp_table[which(tmp_table$cov <= mean(tmp_table$cov)+sd(tmp_table$cov)), ]
coverage_table = rbind(coverage_table, tmp_table)
CNV_table[which(CNV_table$strain == strain_name), "stop_y"] = max(tmp_table$cov)
}
#------------------------------------------------------------------------------#
## 4.2 - plot coverage ####
# format
coverage_table$chr = factor(
coverage_table$chr,
levels = c("I", "II", "III", "IV", "V", "VI", "VII", "VIII",
"IX", "X", "XI", "XII", "XIII", "XIV", "XV", "XVI")
)
CNV_table$chr = factor(
CNV_table$chr,
levels = c("I", "II", "III", "IV", "V", "VI", "VII", "VIII",
"IX", "X", "XI", "XII", "XIII", "XIV", "XV", "XVI")
)
for(k in 1:nrow(to_swap)){
coverage_table$strain = stringr::str_replace_all(
coverage_table$strain,
to_swap[k, "old_name"],
to_swap[k, "new_name"]
)
CNV_table$strain = stringr::str_replace_all(
CNV_table$strain,
to_swap[k, "old_name"],
to_swap[k, "new_name"]
)
average_table$strain = stringr::str_replace_all(
average_table$strain,
to_swap[k, "old_name"],
to_swap[k, "new_name"]
)
}
# relevel
coverage_table$strain = factor(coverage_table$strain, levels = rev(tip_order))
CNV_table$strain = factor(CNV_table$strain, levels = rev(tip_order))
average_table$strain = factor(average_table$strain, levels = rev(tip_order))
# debug
# average_table = average_table[which(average_table$strain == "Voss1"), ]
# coverage_table = coverage_table[which(coverage_table$strain == "Voss1"), ]
# CNV_table = CNV_table[which(CNV_table$strain == "Voss1"), ]
# plot
p_CNV = ggplot() +
geom_rect(data = subset(
coverage_table,
chr %in% c("I", "III", "V", "VII", "IX", "XI", "XIII", "XV")),
fill = "grey95",
xmin = 0, xmax = max(coverage_table$stop),
ymin = 0, ymax = max(coverage_table$cov),
alpha = 0.3) +
geom_rect(data = CNV_table,
aes(fill = CNV,
xmin = start, xmax = stop,
ymin = start_y, ymax = stop_y),
alpha = 0.75) +
geom_hline(data = average_table,
aes(yintercept = average),
color = "firebrick",
linewidth = 1.5) +
geom_point(data = coverage_table,
aes(x = start, y = cov),
size = 0.05, color = "grey35", alpha = 0.5) +
scale_fill_manual(values = c("steelblue", "white", "salmon", "grey95")) +
scale_y_continuous(labels = function(x) format(x, big.mark = ",", scientific = FALSE)) +
coord_cartesian(expand = FALSE) +
annotation_custom(grid::linesGrob(y = c(0, 0), gp = grid::gpar(lwd = 3))) +
facet_grid(strain ~ chr,
scales = "free",
space = "free_x",
switch = "both") +
theme(plot.title = element_blank(),
axis.title = element_blank(),
axis.ticks = element_blank(),
axis.text = element_blank(),
legend.position = "none",
panel.background = element_blank(),
panel.border = element_blank(),
panel.spacing = unit(0, "lines"),
strip.background = element_blank(),
strip.text.x = element_text(size = 8),
strip.text.y.left = element_blank())18.1.2 Merge
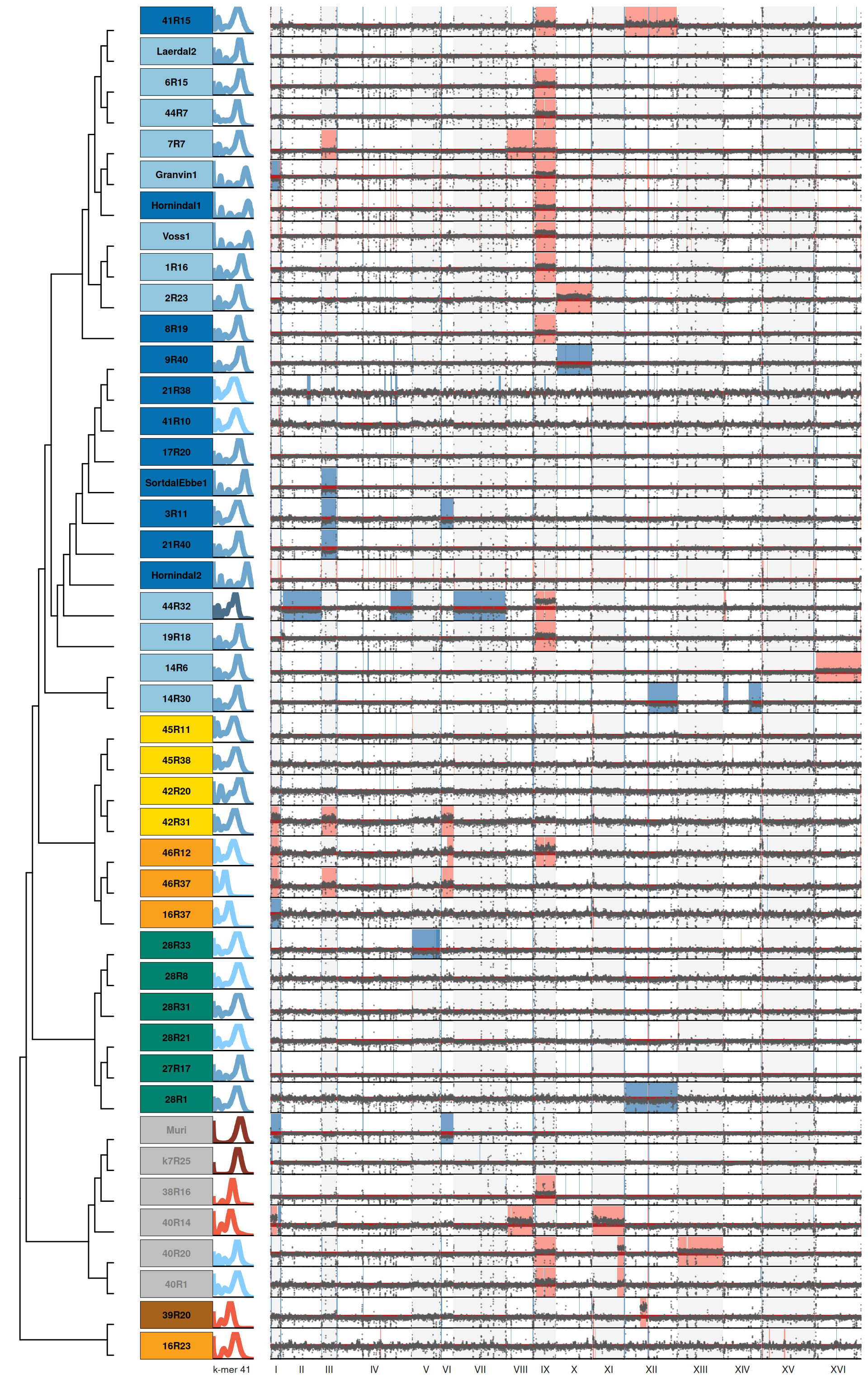
18.2 Figure S6 plot
18.3 Session Information
Note
R version 4.3.3 (2024-02-29)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 24.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.12.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=en_US.UTF-8 LC_MONETARY=C LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
time zone: Europe/Brussels
tzcode source: system (glibc)
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] treeio_1.26.0 stringr_1.5.1 rstatix_0.7.2
[4] ReactomePA_1.46.0 RColorBrewer_1.1-3 plyr_1.8.9
[7] ggtreeExtra_1.12.0 ggtree_3.10.1 ggnewscale_0.5.2
[10] gggenomes_1.0.1 ggh4x_0.3.1 gridExtra_2.3
[13] enrichplot_1.22.0 EnhancedVolcano_1.20.0 ggrepel_0.9.6
[16] ggplot2_3.5.2 edgeR_4.0.16 limma_3.58.1
[19] dplyr_1.1.4 DOSE_3.28.2 ComplexHeatmap_2.18.0
[22] colorRamp2_0.1.0 clusterProfiler_4.10.1 BiocManager_1.30.26
[25] aplot_0.2.8 ape_5.8-1
loaded via a namespace (and not attached):
[1] jsonlite_2.0.0 shape_1.4.6.1 magrittr_2.0.3
[4] farver_2.1.2 rmarkdown_2.29 GlobalOptions_0.1.2
[7] fs_1.6.6 zlibbioc_1.48.2 vctrs_0.6.5
[10] memoise_2.0.1 RCurl_1.98-1.17 htmltools_0.5.8.1
[13] broom_1.0.9 Formula_1.2-5 gridGraphics_0.5-1
[16] htmlwidgets_1.6.4 cachem_1.1.0 igraph_2.1.4
[19] lifecycle_1.0.4 iterators_1.0.14 pkgconfig_2.0.3
[22] Matrix_1.6-5 R6_2.6.1 fastmap_1.2.0
[25] gson_0.1.0 GenomeInfoDbData_1.2.11 clue_0.3-66
[28] digest_0.6.37 colorspace_2.1-1 patchwork_1.3.1
[31] AnnotationDbi_1.64.1 S4Vectors_0.40.2 RSQLite_2.4.2
[34] ggpubr_0.6.1 labeling_0.4.3 abind_1.4-8
[37] httr_1.4.7 polyclip_1.10-7 compiler_4.3.3
[40] bit64_4.6.0-1 withr_3.0.2 doParallel_1.0.17
[43] backports_1.5.0 graphite_1.48.0 S7_0.2.0
[46] BiocParallel_1.36.0 carData_3.0-5 viridis_0.6.5
[49] DBI_1.2.3 ggforce_0.5.0 ggsignif_0.6.4
[52] MASS_7.3-60.0.1 rappdirs_0.3.3 rjson_0.2.23
[55] HDO.db_0.99.1 tools_4.3.3 scatterpie_0.2.5
[58] glue_1.8.0 nlme_3.1-164 GOSemSim_2.28.1
[61] shadowtext_0.1.5 cluster_2.1.6 reshape2_1.4.4
[64] fgsea_1.35.6 generics_0.1.4 gtable_0.3.6
[67] tzdb_0.5.0 tidyr_1.3.1 hms_1.1.3
[70] data.table_1.17.8 car_3.1-3 tidygraph_1.3.1
[73] XVector_0.42.0 BiocGenerics_0.48.1 foreach_1.5.2
[76] pillar_1.11.0 yulab.utils_0.2.0 circlize_0.4.16
[79] splines_4.3.3 tweenr_2.0.3 lattice_0.22-5
[82] bit_4.6.0 tidyselect_1.2.1 GO.db_3.18.0
[85] locfit_1.5-9.12 Biostrings_2.70.3 reactome.db_1.86.2
[88] knitr_1.50 IRanges_2.36.0 stats4_4.3.3
[91] xfun_0.52 graphlayouts_1.2.2 Biobase_2.62.0
[94] statmod_1.5.0 matrixStats_1.5.0 stringi_1.8.7
[97] lazyeval_0.2.2 ggfun_0.2.0 yaml_2.3.10
[100] evaluate_1.0.4 codetools_0.2-19 ggraph_2.2.1
[103] tibble_3.3.0 qvalue_2.34.0 graph_1.80.0
[106] ggplotify_0.1.2 cli_3.6.5 dichromat_2.0-0.1
[109] Rcpp_1.1.0 GenomeInfoDb_1.38.8 png_0.1-8
[112] parallel_4.3.3 ellipsis_0.3.2 readr_2.1.5
[115] blob_1.2.4 bitops_1.0-9 viridisLite_0.4.2
[118] tidytree_0.4.6 scales_1.4.0 purrr_1.1.0
[121] crayon_1.5.3 GetoptLong_1.0.5 rlang_1.1.6
[124] cowplot_1.2.0 fastmatch_1.1-6 KEGGREST_1.42.0