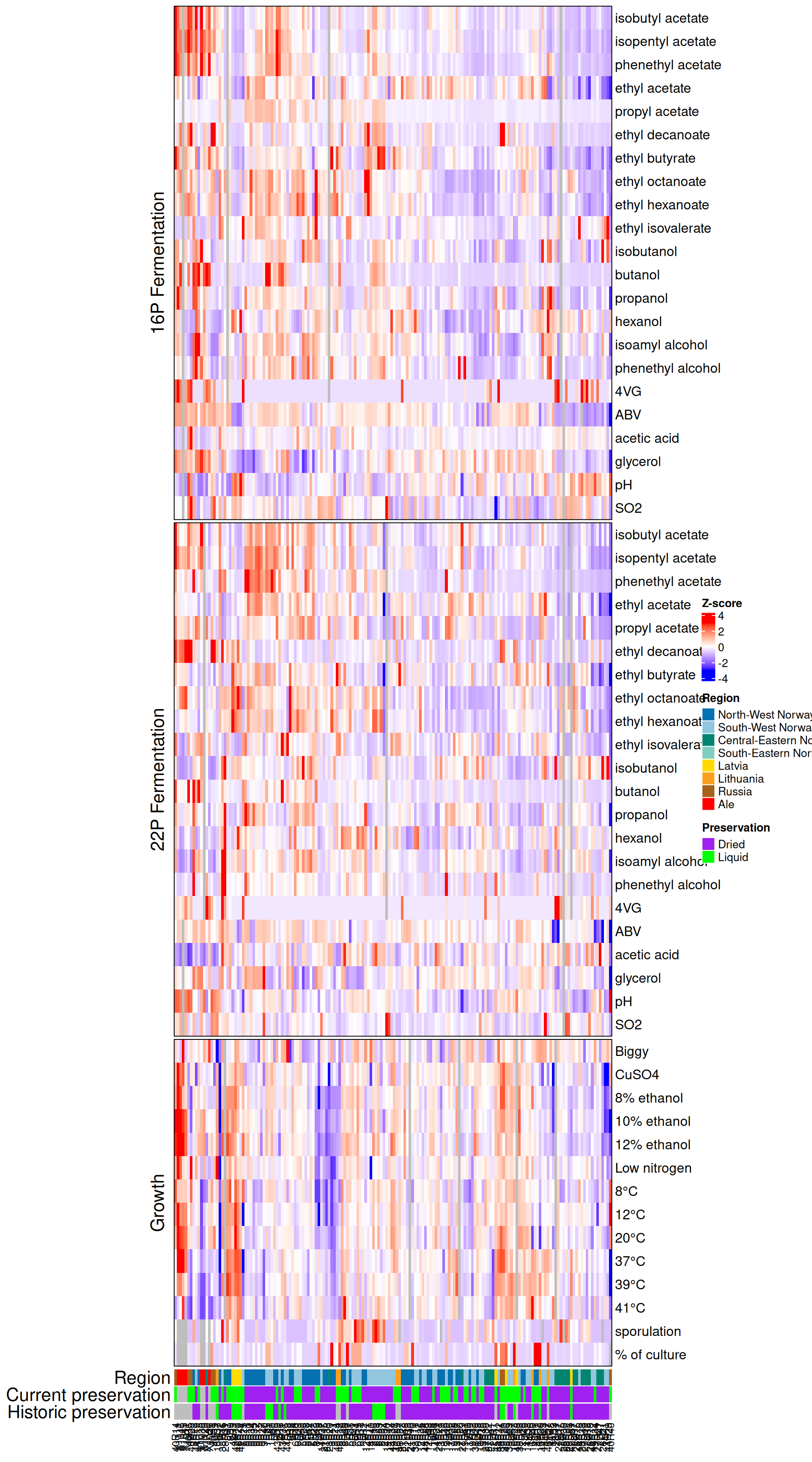
12 Figure 6
12.1 Figure 6 code
12.1.1 Panel A code
################################################################################
# Load fermentation data and set up color functions
################################################################################
label.colours <- brewer.pal(7, 'Paired')
col_fun <- colorRamp2(c(-3, 0, 3), c("blue", "white", "red"))
compound_split <- factor(c(rep("16P Fermentation", 22),
rep("22P Fermentation", 22),
rep("Growth", 14)),
levels = c("16P Fermentation", "22P Fermentation", "Growth"))
mydata <- read.csv('./data/p02-06/data for R FILTERED.csv')
annotations <- mydata[1:4]
row_split <- factor(annotations$region, levels = c("Ale",
"Russia",
"Latvia",
"Lithuania",
"Central-Eastern Norway",
"North-West Norway",
"South-West Norway"))
rownames(mydata) <- mydata[,1]
mydata <- mydata[,-c(1:4)]
mydata <- as.matrix(mydata)
mydata <- t(mydata) # Transpose the matrix
################################################################################
# Create annotations with legends disabled
################################################################################
# For Historic_preservation, use anno_simple so it appears without generating a legend.
ha <- HeatmapAnnotation(
Region = annotations$region,
`Current preservation` = annotations$current_preservation,
`Historic preservation` = annotations$historic_preservation,
col = list(
Region = c('North-West Norway' = '#0571B0',
'South-West Norway' = '#92C5DE',
'Central-Eastern Norway' = '#018571',
'South-Eastern Norway' = '#80CDC1',
'Latvia' = '#FFDA00',
'Lithuania' = '#FBA01D',
'Russia' = '#A6611A',
'Ale' = '#FF0000'),
`Current preservation` = c('Dried' = 'purple', 'Liquid' = 'green'),
`Historic preservation` = c('Dried' = 'purple', 'Liquid' = 'green')
),
# Disable legends for vector annotations
annotation_legend_param = list(
Region = list(legend = FALSE),
`Current preservation` = list(legend = FALSE),
`Historic preservation` = list(legend = FALSE)
),
annotation_name_side = "left",
annotation_name_gp = gpar(fontsize = 16)
)
################################################################################
# Create heatmap with default legends disabled
################################################################################
# Assume mydata already has unique rownames
custom_labels <- c("isobutyl acetate",
"isopentyl acetate",
"phenethyl acetate",
"ethyl acetate",
"propyl acetate",
"ethyl decanoate",
"ethyl butyrate",
"ethyl octanoate",
"ethyl hexanoate",
"ethyl isovalerate",
"isobutanol",
"butanol",
"propanol",
"hexanol",
"isoamyl alcohol",
"phenethyl alcohol",
"4VG",
"ABV",
"acetic acid",
"glycerol",
"pH",
"SO2",
"isobutyl acetate",
"isopentyl acetate",
"phenethyl acetate",
"ethyl acetate",
"propyl acetate",
"ethyl decanoate",
"ethyl butyrate",
"ethyl octanoate",
"ethyl hexanoate",
"ethyl isovalerate",
"isobutanol",
"butanol",
"propanol",
"hexanol",
"isoamyl alcohol",
"phenethyl alcohol",
"4VG",
"ABV",
"acetic acid",
"glycerol",
"pH",
"SO2",
"Biggy",
"CuSO4",
"8% ethanol",
"10% ethanol",
"12% ethanol",
"Low nitrogen",
"8°C",
"12°C",
"20°C",
"37°C",
"39°C",
"41°C",
"sporulation",
"% of culture")
hm <- Heatmap(mydata,
col = col_fun,
border = TRUE,
name = 'Z-score',
cluster_rows = FALSE,
cluster_columns = TRUE,
cluster_column_slices = FALSE,
cluster_row_slices = FALSE,
show_row_dend = FALSE,
show_column_dend = FALSE,
row_split = compound_split,
row_labels = custom_labels,
row_title_gp = gpar(fontsize = 16),
clustering_distance_columns = "manhattan",
right_annotation = NULL,
bottom_annotation = ha,
column_names_gp = grid::gpar(fontsize = 9),
column_title = NULL,
show_heatmap_legend = FALSE)
################################################################################
# Manually create legend objects
################################################################################
# Heatmap (Z-score) legend
lg_heatmap <- Legend(col_fun = col_fun, title = "Z-score")
# Region legend (adjust levels and colors as needed)
lg_region <- Legend(title = "Region",
at = c("North-West Norway", "South-West Norway", "Central-Eastern Norway",
"South-Eastern Norway", "Latvia", "Lithuania", "Russia", "Ale"),
legend_gp = gpar(fill = c("#0571B0", "#92C5DE", "#018571", "#80CDC1",
"#FFDA00", "#FBA01D", "#A6611A", "#FF0000")))
# Preservation legend: combining both preservation annotations
lg_preservation <- Legend(title = "Preservation",
at = c("Dried", "Liquid"),
legend_gp = gpar(fill = c("purple", "green")))
################################################################################
# Pack legends vertically
################################################################################
combined_legends <- packLegend(lg_heatmap, lg_region, lg_preservation, direction = "vertical")12.2 Figure 6 plot
12.3 Session Information
Note
R version 4.3.3 (2024-02-29)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 24.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.12.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=en_US.UTF-8 LC_MONETARY=C LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
time zone: Europe/Brussels
tzcode source: system (glibc)
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] vegan_2.7-1 permute_0.9-8 umap_0.2.10.0
[4] RColorBrewer_1.1-3 patchwork_1.3.1 ComplexHeatmap_2.18.0
[7] circlize_0.4.16
loaded via a namespace (and not attached):
[1] generics_0.1.4 shape_1.4.6.1 lattice_0.22-5
[4] digest_0.6.37 magrittr_2.0.3 evaluate_1.0.4
[7] iterators_1.0.14 fastmap_1.2.0 Matrix_1.6-5
[10] foreach_1.5.2 doParallel_1.0.17 jsonlite_2.0.0
[13] RSpectra_0.16-2 GlobalOptions_0.1.2 mgcv_1.9-1
[16] scales_1.4.0 codetools_0.2-19 cli_3.6.5
[19] rlang_1.1.6 crayon_1.5.3 splines_4.3.3
[22] yaml_2.3.10 tools_4.3.3 parallel_4.3.3
[25] dplyr_1.1.4 colorspace_2.1-1 ggplot2_3.5.2
[28] GetoptLong_1.0.5 BiocGenerics_0.48.1 reticulate_1.43.0
[31] vctrs_0.6.5 R6_2.6.1 png_0.1-8
[34] magick_2.8.7 matrixStats_1.5.0 stats4_4.3.3
[37] lifecycle_1.0.4 S4Vectors_0.40.2 htmlwidgets_1.6.4
[40] IRanges_2.36.0 MASS_7.3-60.0.1 clue_0.3-66
[43] cluster_2.1.6 pkgconfig_2.0.3 pillar_1.11.0
[46] gtable_0.3.6 glue_1.8.0 Rcpp_1.1.0
[49] xfun_0.52 tibble_3.3.0 tidyselect_1.2.1
[52] knitr_1.50 dichromat_2.0-0.1 farver_2.1.2
[55] rjson_0.2.23 nlme_3.1-164 htmltools_0.5.8.1
[58] rmarkdown_2.29 compiler_4.3.3 askpass_1.2.1
[61] openssl_2.3.3